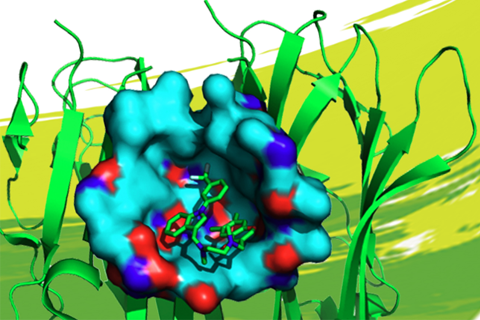
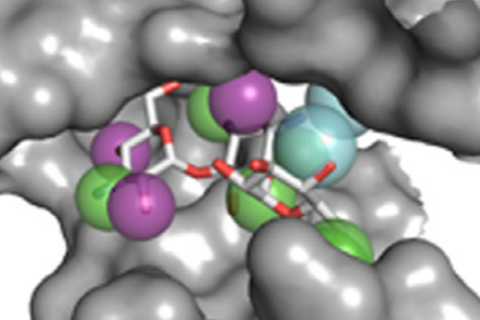
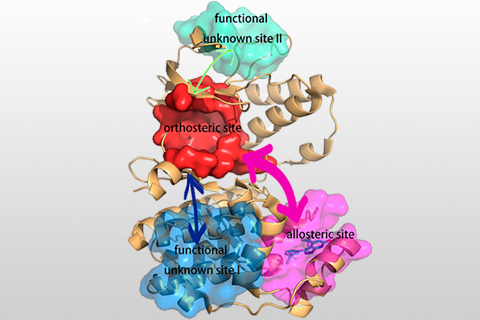
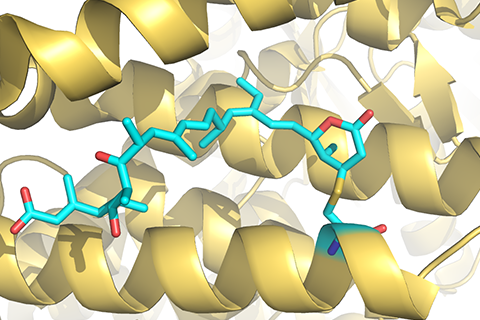
Identifying reliable binding sites based on three dimensional structures of proteins and other macromolecules is a key step in drug discovery. A good definition of known binding site and the detection of a novel site can provide valuable information for drug design efforts. CAVITY is developed for the detection and analysis of ligand-binding site(s) . It has the capability of detecting potential binding site as well as estimating both the ligandabilities and druggabilites of the detected binding sites.
CAVITY was originally used in the de novo drug design tool LigBuilder 2.0 to accurately reflect the key interactions within a binding site as well as to confine the ligand growth within a reasonable region; it was later developed into a stand-alone program for binding site detection and analysis. The CAVITY approach generates clear and accurate information about the shapes and boundaries of the ligand binding sites, which provide helpful information for drug discovery studies: 1) For cases where a protein-ligand complex of the target protein is available, CAVITY can be used to detect the binding site regions which are not covered by the known ligand(s) and provide clues for the improvement of ligand-binding affinity. In addition, the predicted ligandability and druggability of the binding site would tell the researchers whether further improvement of the known ligand is promising. 2) For cases where ligands are known, but the structural information of ligand-target interactions is not available, CAVITY can be used to detect the binding site and the binding mode of the known ligands could be predicted by using molecular docking technique. 3) For cases with no reported ligand, CAVITY can not only be used to detect potential binding sites, but also to provide qualitative estimations of ligandability and druggability for potential binding sites on the target protein, which is very important for making an early stage decision on whether the protein is a promising target for a drug discovery project. CAVITY has been used in many different projects to help generate such information and clues. We used the external NRDLD data set and Cheng’s data set to test Cavity, showing a satisfactory performance of 0.82 and 0.89 accuracy, respectively.
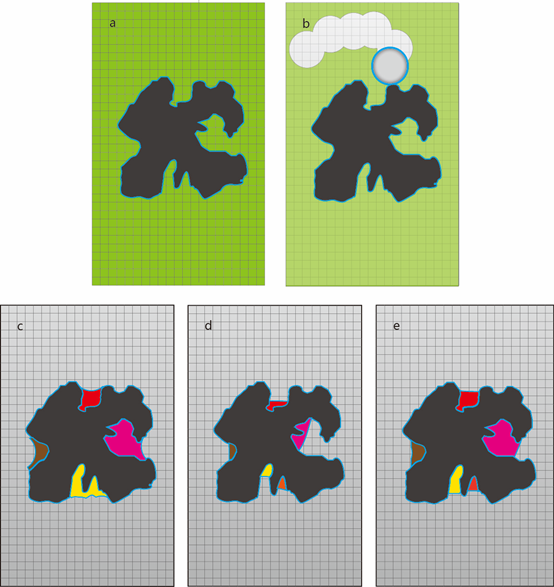
Schematic diagram of cavity detection in CAVITY. a) Protein (black-colored) in grid box (green-colored). b) Using the eraser ball to remove grid points outside protein. c) "Vacant" grid points after erasing. Four cavities were shown in different colors. d) Shrink each cavity until the depth reach the minimal depth. e) Recover cavities to obtain the final result.